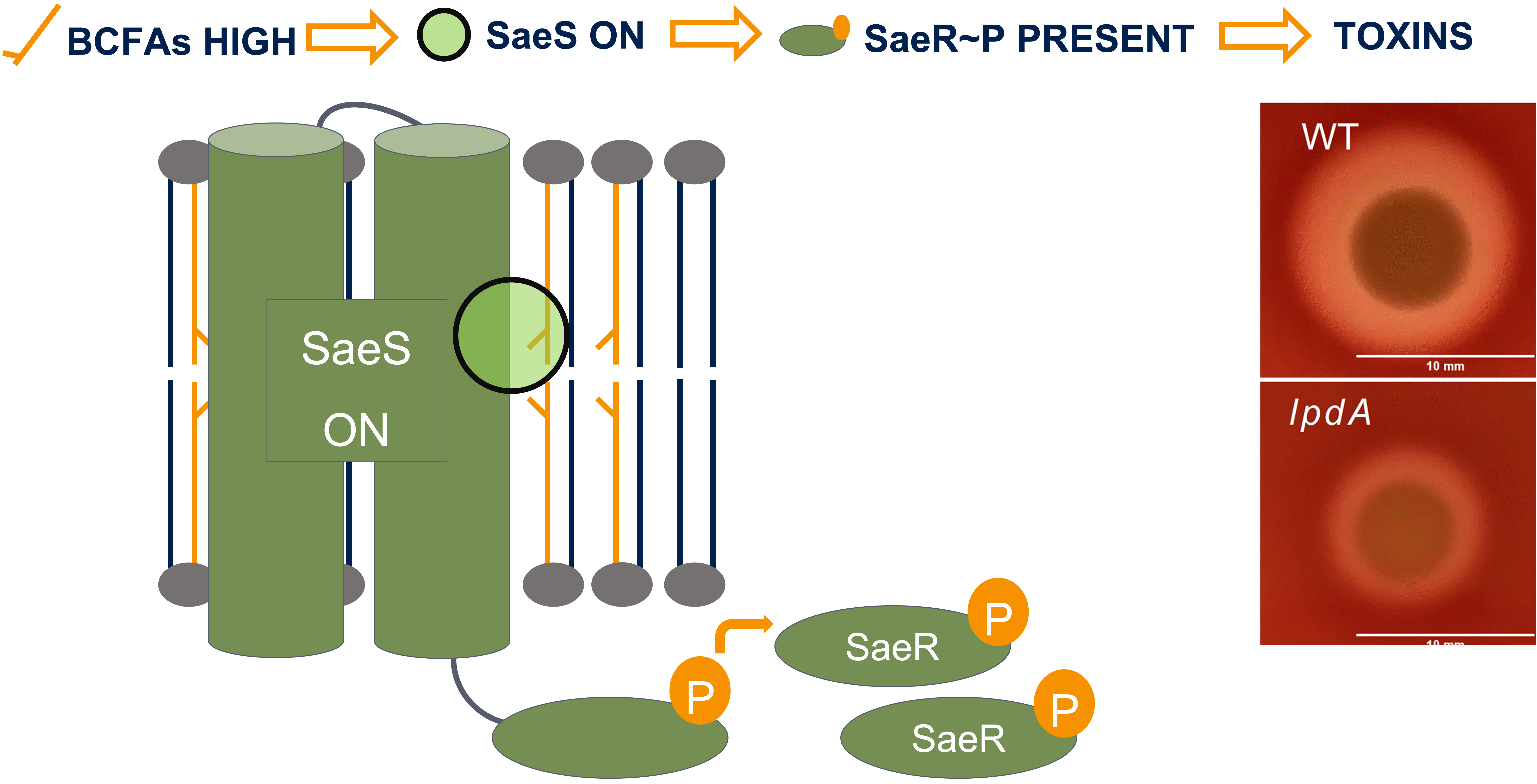
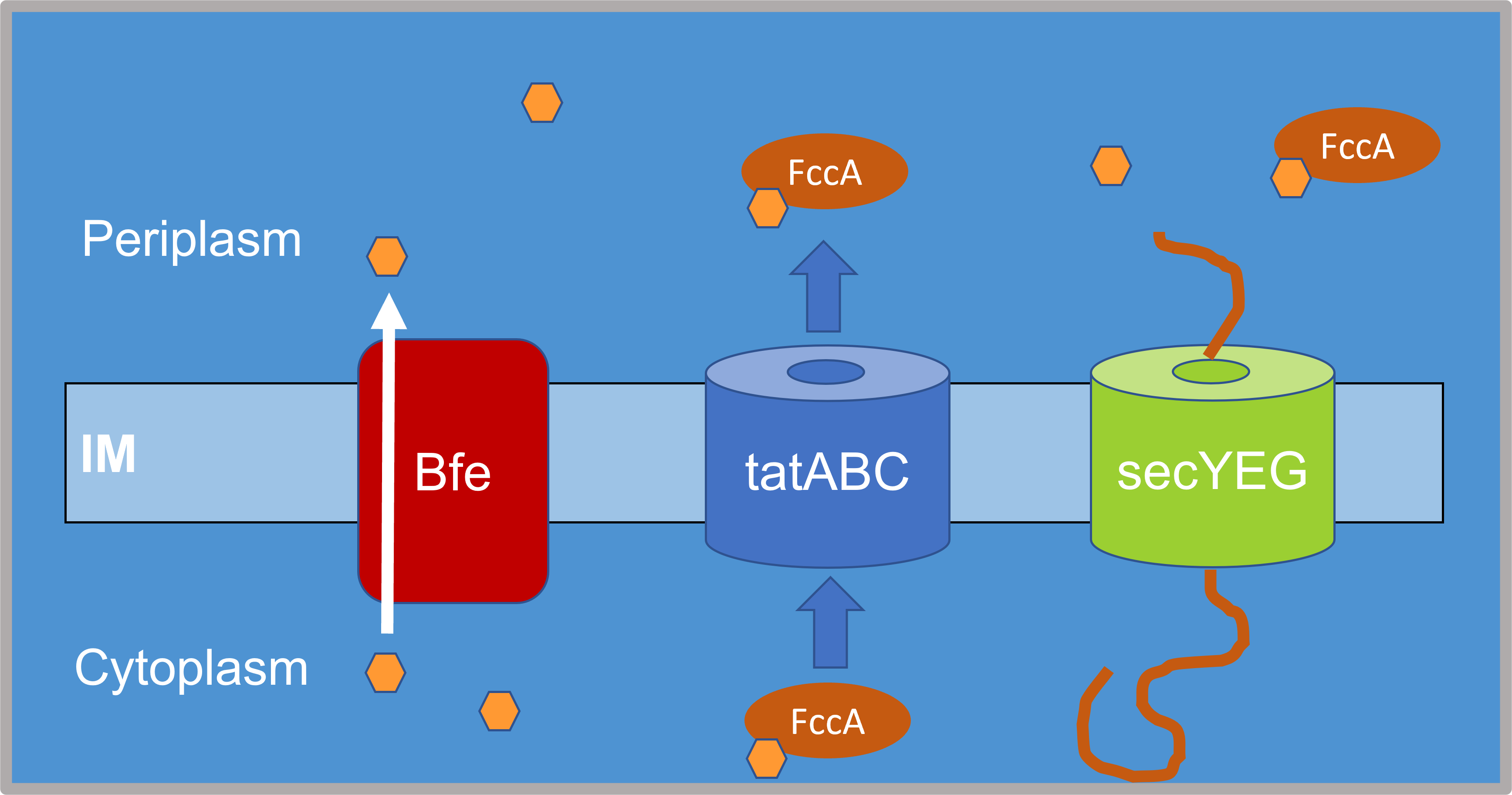
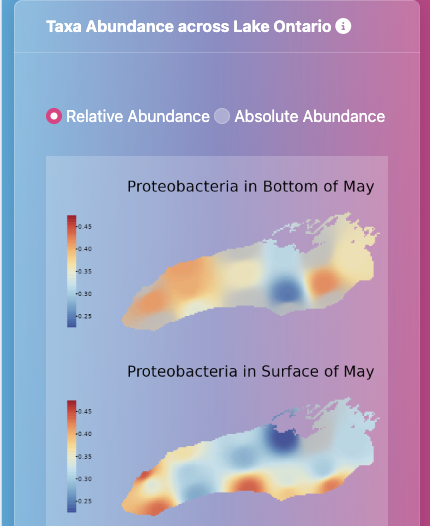
Microbes are the foundation of healthy freshwater ecosystems and their potential to remediate environmental damage drives my interest in microbial ecology. I use fieldwork, molecular biology, and bioinformatics to study the bacteria that live in freshwater ecosystems, including the Great Lakes (via the EPA CSMI cruises), the Lake Superior National Estuarine Research Reserve (through a NOAA Davidson Fellowship), and at Cornell's own experimental ponds. I study how physical processes (like stratification and circulation) affect the ecological processes of selection, dispersal, and drift in microbial populations and communities. I'm also an R and Julia programmer and teach coding through the Software Carpentries. On this page you can access my publications and some of my personal projects as a bioinformatician. You can also view my CV here. Always happy to chat about bacteria, coding, or freshwater, so please feel free to reach out via BlueSky or email.